Plotting Tutorial¶
General Tips¶
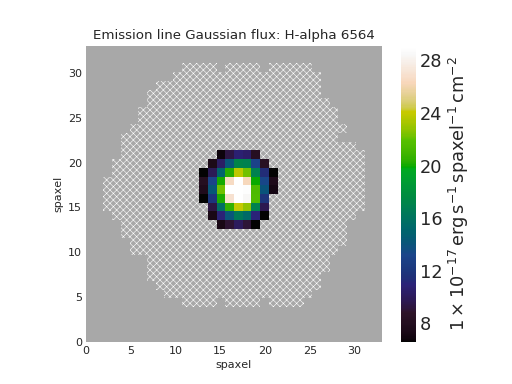
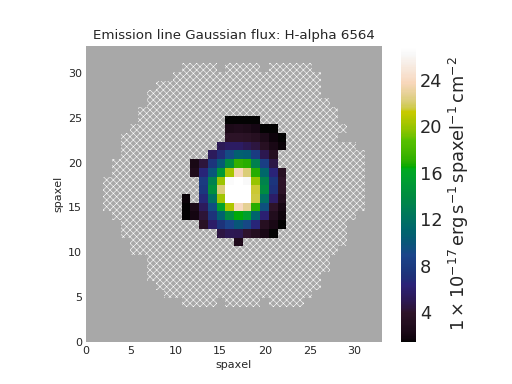
Quick Map Plot¶
from marvin.tools import Maps
maps = Maps('8485-1901')
ha = maps.emline_gflux_ha_6564
ha.plot()
(Source code, png, hires.png, pdf)
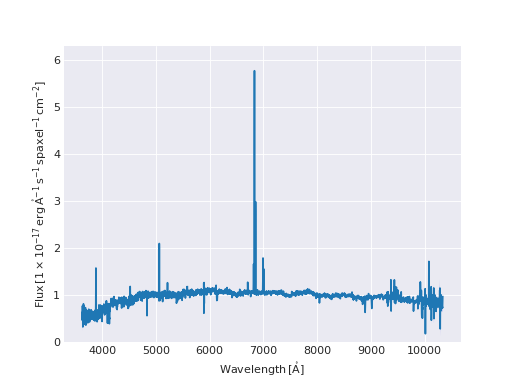
Quick Spectrum Plot¶
from marvin.tools import Cube
cube = Cube('8485-1901')
spax = cube[17, 17]
spax.flux.plot()
(Source code, png, hires.png, pdf)
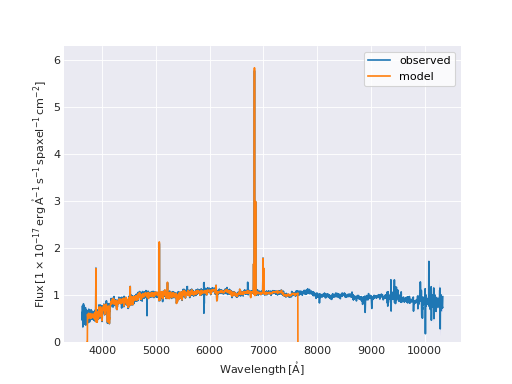
Quick Model Fit Plot¶
from marvin.tools import Maps
maps = Maps('8485-1901')
# must use Maps.getSpaxel() to get cube and modelcube
spax = maps.getSpaxel(x=17, y=17, xyorig='lower', cube=True, modelcube=True)
# mask out pixels lacking model fit
no_fit = ~spax.full_fit.masked.mask
# extra arguments to plot are passed to the matplotlib routine
ax = spax.flux.plot(label='observed')
ax.plot(spax.full_fit.wavelength[no_fit], spax.full_fit.value[no_fit], label='model')
ax.legend()
(Source code, png, hires.png, pdf)
Quick Image Plot¶
import matplotlib.pyplot as plt
from marvin.tools.image import Image
image = Image(plateifu='8553-12702')
image.plot()
(Source code, png, hires.png, pdf)
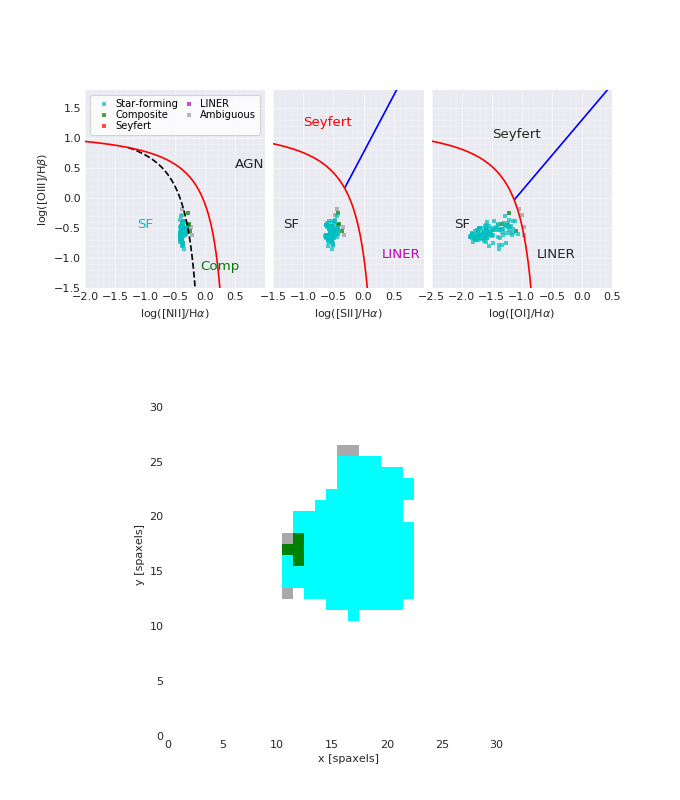
BPT Plot¶
from marvin.tools import Maps
maps = Maps('8485-1901')
masks, fig, axes = maps.get_bpt()
(Source code, png, hires.png, pdf)
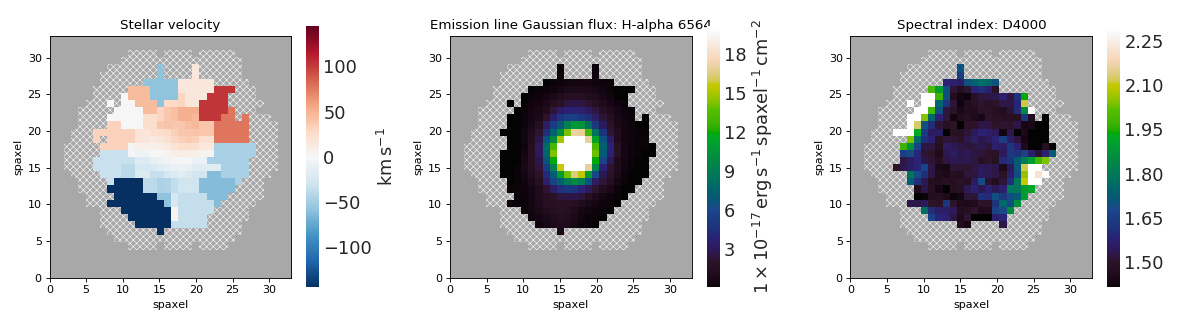
Multi-panel Map Plot (Single Galaxy)¶
import matplotlib.pyplot as plt
from marvin.tools import Maps
import marvin.utils.plot.map as mapplot
maps = Maps('8485-1901')
stvel = maps['stellar_vel']
ha = maps['emline_gflux_ha_6564']
d4000 = maps['specindex_d4000']
fig, axes = plt.subplots(1, 3, figsize=(15, 4))
for ax, map_ in zip(axes, [stvel, ha, d4000]):
mapplot.plot(dapmap=map_, fig=fig, ax=ax)
fig.tight_layout()
(Source code, png, hires.png, pdf)
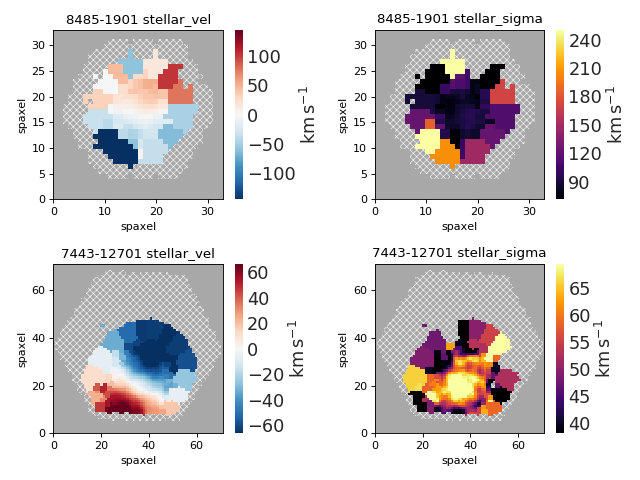
Multi-panel Map Plot (Multiple Galaxies)¶
import matplotlib.pyplot as plt
from marvin.tools import Maps
import marvin.utils.plot.map as mapplot
plateifus = ['8485-1901', '7443-12701']
mapnames = ['stellar_vel', 'stellar_sigma']
rows = len(plateifus)
cols = len(mapnames)
fig, axes = plt.subplots(rows, cols, figsize=(8, 6))
for row, plateifu in zip(axes, plateifus):
maps = Maps(plateifu=plateifu)
for ax, mapname in zip(row, mapnames):
mapplot.plot(dapmap=maps[mapname], fig=fig, ax=ax, title=' '.join((plateifu, mapname)))
fig.tight_layout()
(Source code, png, hires.png, pdf)
Zoom-in Map Plot¶
from marvin.tools import Maps
maps = Maps('8485-1901')
ha = maps.emline_gflux_ha_6564
fig, ax = ha.plot()
ax.axis([13, 21, 13, 21])
(Source code, png, hires.png, pdf)
Custom Map Colorbar Range Options¶
:align: center
:include-source: True
from marvin.tools import Maps
maps = Maps('8485-1901')
ha = maps.emline_gflux_ha_6564
fig, ax = ha.plot(percentile_clip=(1, 99))
fig, ax = ha.plot(sigma_clip=2)
fig, ax = ha.plot(cbrange=[2, 10])
fig, ax = ha.plot(symmetric=True)
fig, ax = ha.plot(log_cb=True)
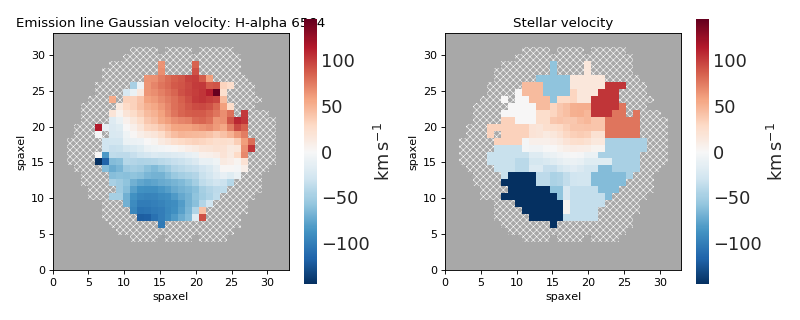
Multi-panel Map Plot with Matching Colorbar Ranges¶
import numpy as np
import matplotlib.pyplot as plt
from marvin.tools import Maps
import marvin.utils.plot.map as mapplot
maps = Maps('8485-1901')
havel = maps.emline_gvel_ha_6564
stvel = maps.stellar_vel
vel_maps = [havel, stvel]
cbranges = [vel_map.plot(return_cbrange=True) for vel_map in vel_maps]
cb_max = np.max(np.abs(cbranges))
cbrange = (-cb_max, cb_max)
fig, axes = plt.subplots(ncols=2, figsize=(10, 4))
for ax, vel_map in zip(axes, vel_maps):
vel_map.plot(fig=fig, ax=ax, cbrange=cbrange)
fig.tight_layout()
(Source code, png, hires.png, pdf)
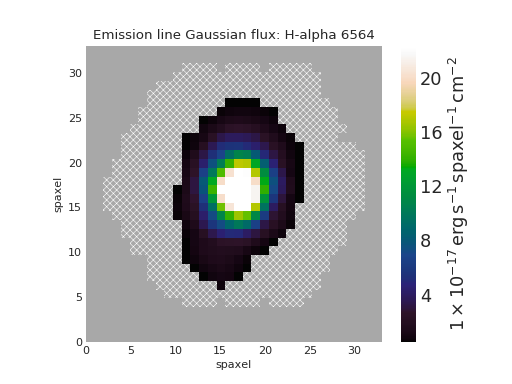
Custom Minimum Signal-to-Noise Ratio¶
from marvin.tools import Maps
maps = Maps('8485-1901')
ha = maps.emline_gflux_ha_6564
# Default is 1 except for velocities, which default to 0.
fig, ax = ha.plot(snr_min=10)
(Source code, png, hires.png, pdf)
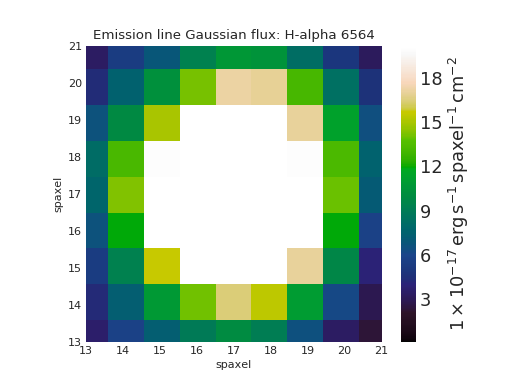
Custom No Usable IFU Data Region¶
from marvin.tools import Maps
maps = Maps('8485-1901')
ha = maps.emline_gflux_ha_6564
# Defaults:
# gray background (facecolor=''#A8A8A8'),
# white lines (edgecolor='w'),
# dense hatching: (hatch= 'xxxx')
# Custom: black background, cyan lines, less dense hatching
fig, ax = ha.plot(patch_kws={'facecolor': 'k',
'edgecolor': 'c',
'hatch': 'xx'})
(Source code, png, hires.png, pdf)
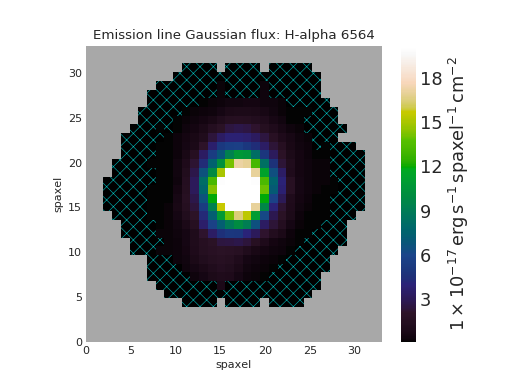
Custom Axis and Colorbar Locations for Map Plot¶
import matplotlib.pyplot as plt
from marvin.tools import Maps
maps = Maps('8485-1901')
ha = maps.emline_gflux_ha_6564
fig = plt.figure()
ax = fig.add_axes([0.12, 0.1, 2 / 3., 5 / 6.])
fig, ax = ha.plot(fig=fig, ax=ax, cb_kws={'axloc': [0.8, 0.1, 0.03, 5 / 6.]})
(Source code, png, hires.png, pdf)
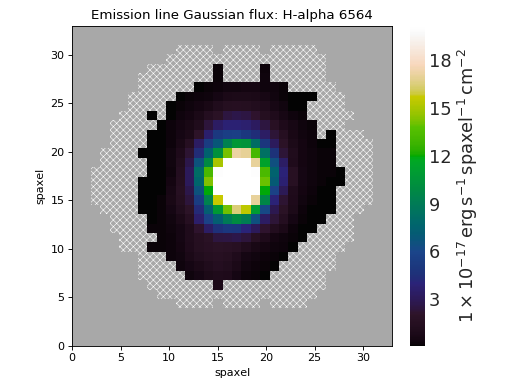
Custom Spectrum and Model Fit¶
import matplotlib.pyplot as plt
from marvin.tools import Maps
plt.style.use('seaborn-darkgrid')
maps = Maps('1-209232')
spax = maps.getSpaxel(x=0, y=0, xyorig='center', cube=True, modelcube=True)
fig, ax = plt.subplots()
pObs = ax.plot(spax.flux.wavelength, spax.flux.value)
pModel = ax.plot(spax.full_fit.wavelength, spax.full_fit.value)
pEmline = ax.plot(spax.emline_fit.wavelength, spax.emline_fit.value)
plt.legend(pObs + pEmline + pModel, ['observed', 'emline model', 'model'])
ax.axis([6700, 7100, -0.1, 3])
ax.set_xlabel('observed wavelength [{}]'.format(spax.flux.wavelength.unit.to_string('latex')))
ax.set_ylabel('flux [{}]'.format(spax.flux.unit.to_string('latex')))
(Source code, png, hires.png, pdf)
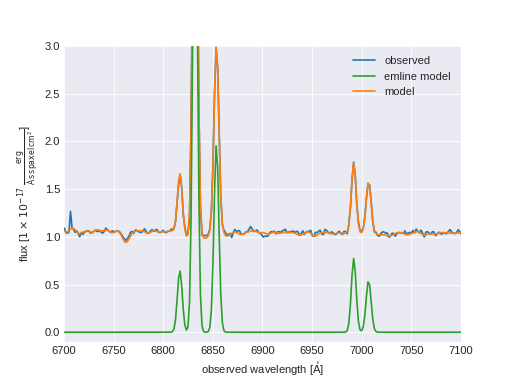
Plot H\(\alpha\) Map of Star-forming Spaxels¶
import numpy as np
from marvin.tools import Maps
maps = Maps('8485-1901')
ha = maps.emline_gflux_ha_6564
masks = maps.get_bpt(show_plot=False, return_figure=False)
# Create a bitmask for non-star-forming spaxels by taking the
# complement (`~`) of the BPT global star-forming mask (where True == star-forming)
# and set bit 30 (DONOTUSE) for those spaxels.
mask_non_sf = ~masks['sf']['global'] * ha.pixmask.labels_to_value('DONOTUSE')
# Do a bitwise OR between DAP mask and non-star-forming mask.
mask = ha.mask | mask_non_sf
ha.plot(mask=mask)
(Source code, png, hires.png, pdf)
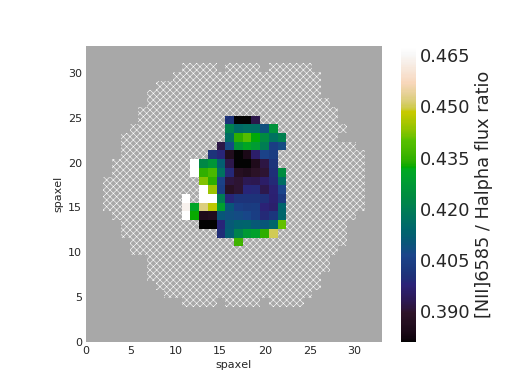
Plot [NII]/H\(\alpha\) Flux Ratio Map of Star-forming Spaxels¶
from marvin.tools import Maps
maps = Maps('8485-1901')
ha = maps.emline_gflux_ha_6564
nii = maps.emline_gflux_nii_6585
nii_ha = nii / ha
# Mask out non-star-forming spaxels
masks, __, __ = maps.get_bpt(show_plot=False)
# Create a bitmask for non-star-forming spaxels by taking the
# complement (`~`) of the BPT global star-forming mask (where True == star-forming)
# and set bit 30 (DONOTUSE) for those spaxels.
mask_non_sf = ~masks['sf']['global'] * ha.pixmask.labels_to_value('DONOTUSE')
# Do a bitwise OR between DAP mask and non-star-forming mask.
mask = nii_ha.mask | mask_non_sf
nii_ha.plot(mask=mask, cblabel='[NII]6585 / Halpha flux ratio')
(Source code, png, hires.png, pdf)
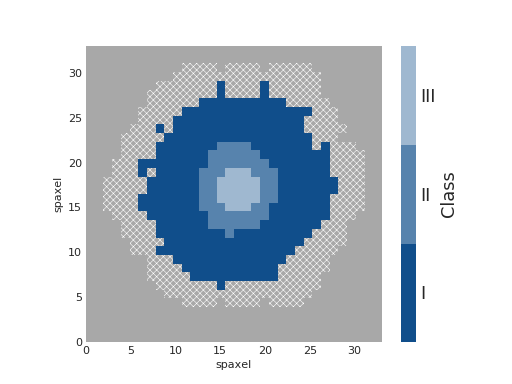
Qualitative Colorbar¶
import numpy as np
from matplotlib import pyplot as plt
from matplotlib.colors import ListedColormap
from marvin.tools import Maps
import marvin.utils.plot.map as mapplot
maps = Maps('8485-1901')
ha = maps.emline_gflux_ha_6564
# divide data into classes
ha_class = np.ones(ha.shape, dtype=int)
ha_class[np.where(ha.value > 5)] = 2
ha_class[np.where(ha.value > 20)] = 3
cmap = ListedColormap(['#104e8b', '#5783ad', '#9fb8d0'])
fig, ax, cb = mapplot.plot(dapmap=ha, value=ha_class, cmap=cmap, cbrange=(0.5, 3.5),
title='', cblabel='Class', return_cb=True)
cb.set_ticks([1, 2, 3])
cb.set_ticklabels(['I', 'II', 'III'])
(Source code, png, hires.png, pdf)
Custom Values and Custom Mask¶
from marvin.tools import Maps
import marvin.utils.plot.map as mapplot
maps = Maps('8485-1901')
ha = maps.emline_gflux_ha_6564
# Mask spaxels without IFU coverage
# nocov = ha.mask & 2**0
nocov = ha.pixmask.get_mask('NOCOV')
# Mask spaxels with low Halpha flux
low_ha = (ha.value < 6) * ha.pixmask.labels_to_value('DONOTUSE')
# Combine masks using bitwise OR (`|`)
mask = nocov | low_ha
fig, ax = mapplot.plot(dapmap=ha, value=ha.value, mask=mask)
(Source code, png, hires.png, pdf)